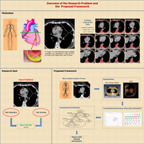
Research Overview:
Cardiovascular Disease (CVD) is one of the leading causes of death both in United States and all around the globe. The National Heart Lung and Blood Institute's 2007 disease statistics reported that (in 2004) 872,000 deaths or 36% of all the deaths in United States were due to cardiovascular disease.
One of the primary causes of CVD is coronary artery atherosclerosis, also popularly known as coronary artery disease (CAD), a condition that refers to a narrowing and hardening of the coronary arteries that supply blood to the heart. This occurs due to atherosclerosis, a disease in which scattered lesions called atherosclerotic plaques (a mixture of fatty substances, cholestrol, cellular wastes, calcium, and the blood clotting material called fibrin) build up on the inner walls of the coronary arteries. Such a condition progressively obstructs the flow of blood through the coronary arteries and when it worsens may eventually lead to a heart attack.
Recent studies have established that the presence of calcified coronary plaques as detected from non-contrast computed tomography (CT) data has a significant predictive value for CAD in both symptomatic and asymptomatic patients. To that end, several risk scores have been developed to quantify the amount of coronary artery calcium (CAC) based on the data collected by CT. However, inspite of the vast amount of CAD-related information available from CT, only a small fraction of it is being used in existing risk scoring strategies. Additionally, most of these scores were devised based on small or diseased populations without rigorous statistical validation. This can be attributed to the lack of robust image analysis methods for the automated extraction of CAD-related information from non-contrast CT imagery. This is precisely the issue that we are trying to address in our ongoing research.
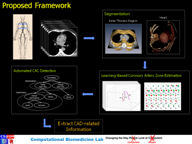
The overall goal of our research is to develop the fundamental set of computational tools that will pave the way for the automated extraction of a variety of CAD-related information from non-contrast CT data.
Below is a list of methods that i developed during the course of my doctoral research:
- A supervised hierarchical classification scheme for CAC detection in non-contrast CT data
- Learning-based estimation of coronary artery zones in non-contrast CT data
- Automatic delineation of the inner-thoracic region in non-contrast CT data
- Automatic segmentation of the diaphragm in non-contrast CT data
- Fuzzy-cuts: Knowledge-driven graph-based method for medical image segmentation
- A shape-driven mrf-model for the segmentation of organs in medical images
- An Explicit Shape-Constrained MRF-based contour evolution method for Image Segmentation
Outreach Material:
Abstract:
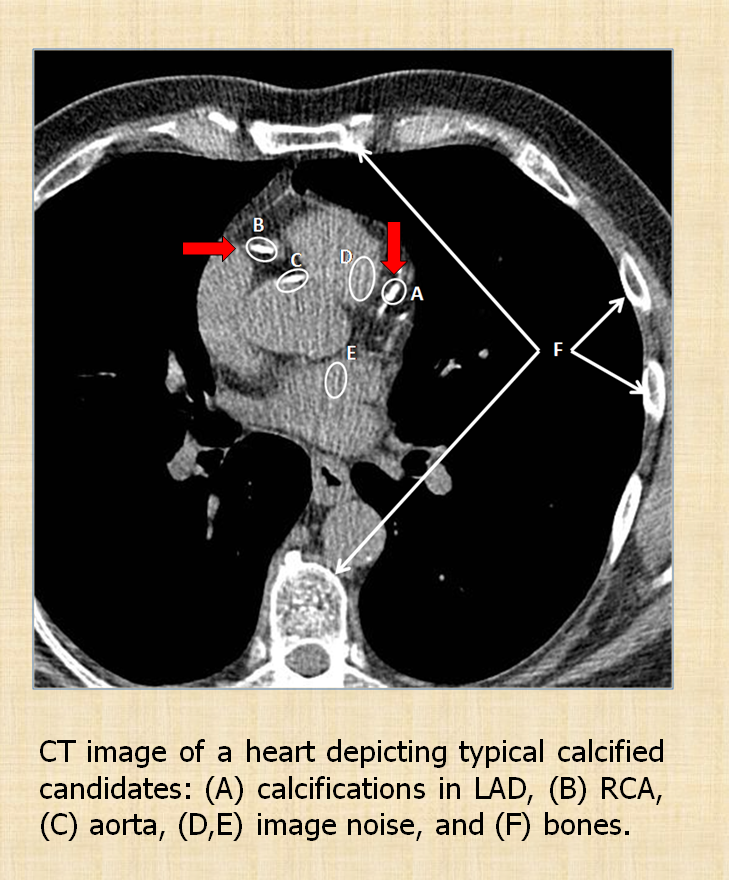
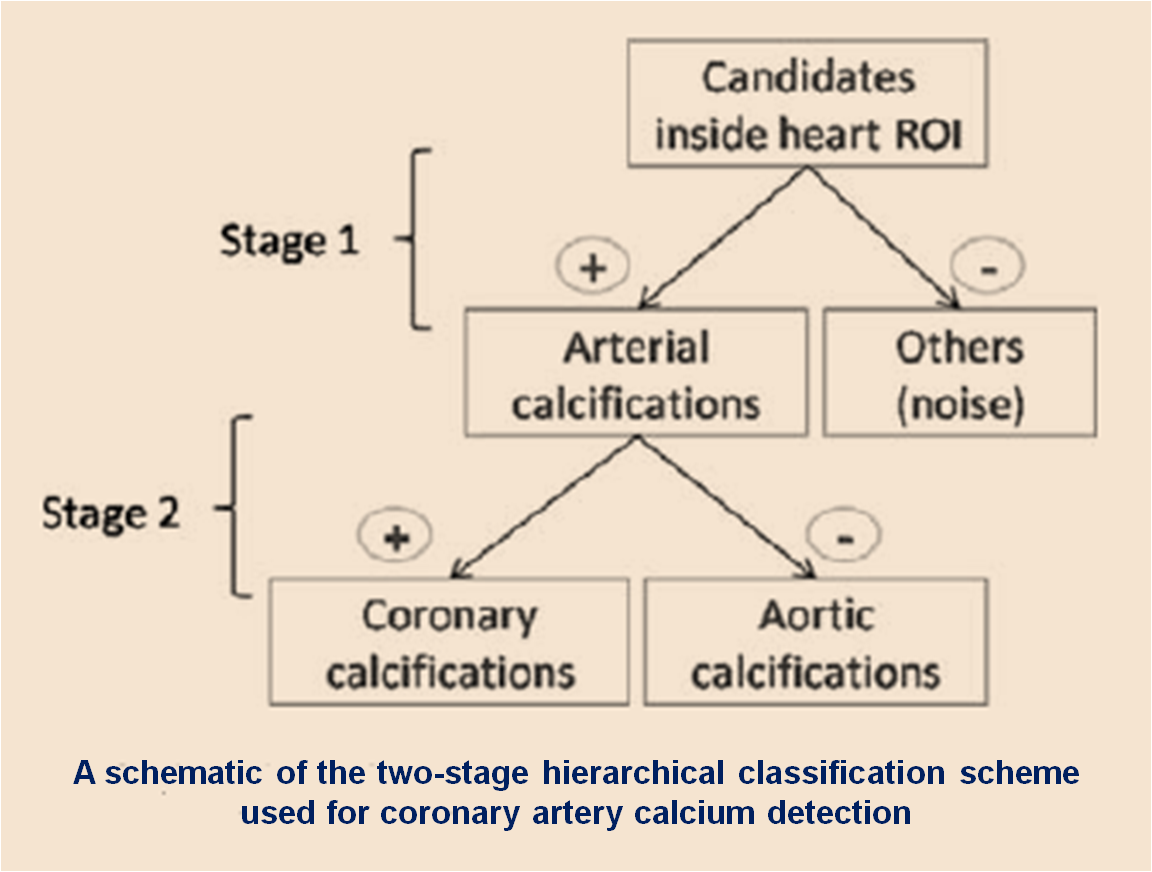
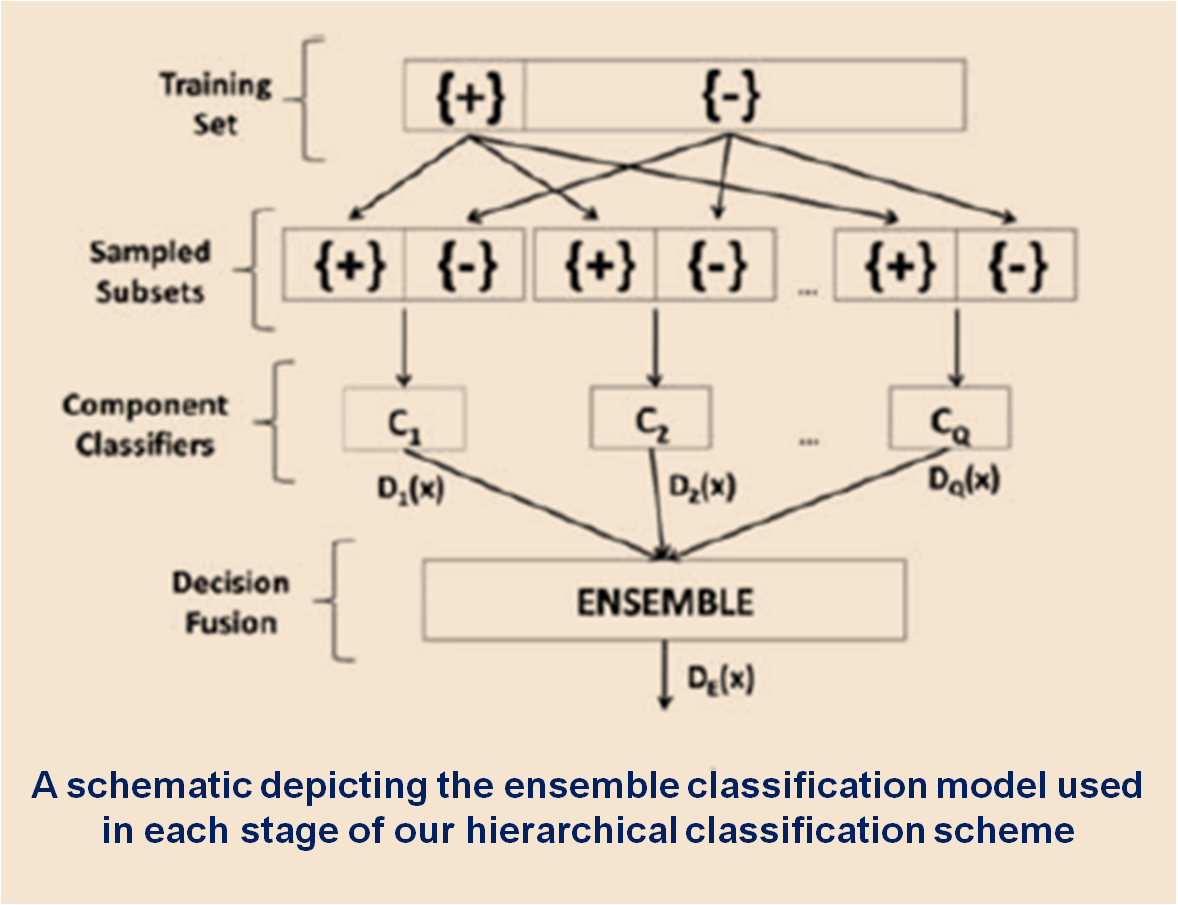
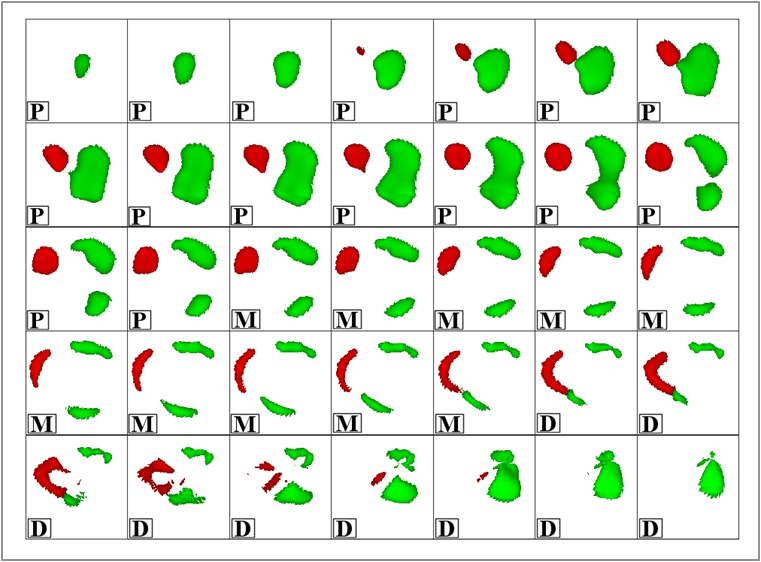
Accurate quantification of coronary artery calcium (CAC) provides an opportunity to assess the extent of atherosclerosis disease. Coronary calcification burden has been reported to be associated with cardiovascular risk. Currently, an observer has to identify the coronary calcifications among a set of candidate regions, obtained by thresholding and connected component labeling, by clicking on them. To relieve the observer of such a labor-intensive task, an automated tool is needed that can detect and quantify the coronary calcifications. However, the diverse and heterogeneous nature of the candidate regions poses a significant challenge. In this paper, we investigate a supervised classification-based approach to distinguish the coronary calcifications from all the candidate regions and propose a two-stage, hierarchical classifier for automated coronary calcium detection. At each stage, we learn an ensemble of classifiers where each classifier is a cost-sensitive learner trained on a distinct asymmetrically sampled data subset. We compute the relative location of the calcifications with respect to a heartcentered coordinate system, and also use the neighboring regions of the calcifications to better characterize their properties for discrimination. Our method detected coronary calcifications with an accuracy, sensitivity and specificity of 98.27, 92.07 and 98.62%, respectively, for a testing dataset of non-contrast computed tomography scans from 105 subjects.
Outreach Material:

Associated Publications:
-
U. Kurkure, D.R. Chittajallu, G. Brunner, R. Yalamanchili, and I.A. Kakadiaris, "Detection of coronary calcifications using supervised hierarchical classication," Proc. of 2nd MICCAI Workshop on Computer Vision for Intravascular and Intracardiac Imaging, New York, NY, Sept. 10, 2008.
-
U. Kurkure, D.R. Chittajallu, G. Brunner, Y.L. Hai, and I.A. Kakadiaris, "A supervised classification-based method for coronary calcium detection in non-contrast CT," International Journal of Cardiovascular Imaging (IJCI), 26(7): 817-28, Oct, 2010.
Abstract:
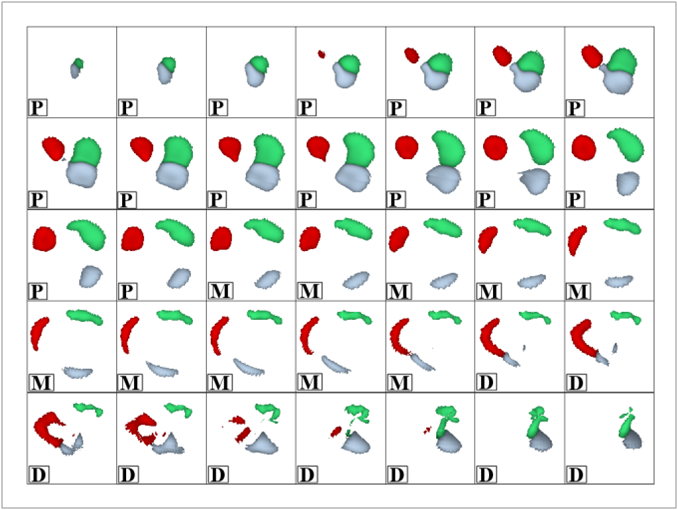
Poor contrast of non-contrast CT scans acquired for calcium scoring makes an accurate segmentation of the coronary arteries an extremely difficult or impossible problem. However, a solution to this problem is very essential for associating the coronary lesions to their location in the coronary artery tree which in turn is of importance for CAD risk prediction. In this work, we present a learning-based method for the estimation of coronary artery zones in non-contrast CT data. We introduce a novel heart-centered coordinate system (HCCS) which is based on anatomical landmarks. The HCCS is used to align non-contrast CT heart volumes for the purpose of developing a supervised classification scheme which discriminates between coronary artery zones and their surrounding tissue. The proposed method has been deployed to 30 non-contrast CT data sets with very encouraging results.
Outreach Material:


Associated Publications:
-
G. Brunner, D.R. Chittajallu, U. Kurkure, I.A. Kakadiaris, "A Heart-centered coordinate system for the detection of coronary artery zones in non-contrast Computed Tomography data," Proc. of 2nd MICCAI Workshop on Computer Vision for Intravascular and Intracardiac Imaging, New York, NY, Sept. 10, 2008.
-
G. Brunner, D.R. Chittajallu, U. Kurkure, and I.A. Kakadiaris, "Toward the automatic detection of coronary artery calcification in non-contrast Computed Tomography data," International Journal of Cardiovascular Imaging, 2009.
Abstract:
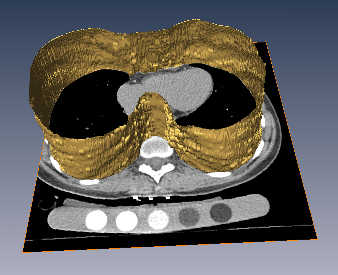
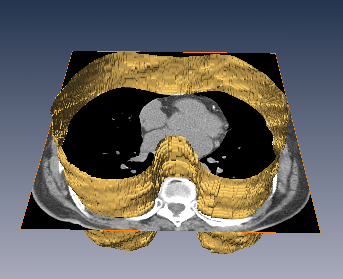
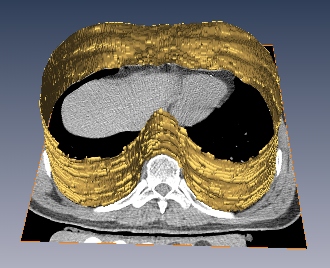
The inner thoracic region consists of several important anatomical structures and an accurate delineation of this region is an essential step for various biomedical image analysis applications. In this work, we present a fully automatic graphbased method for the delineation of the inner thoracic region in non-contrast cardiac CT data. In particular, we reformulate the problem of delineating the inner thoracic region as an optimal surface segmentation problem, the solution to which is obtained by computing the minimum-cost closed set in a node-weighted directed graph. Comparing the results obtained using our method with manual segmentations performed by an expert on non-contrast cardiac CT scans of 20 randomly selected patients indicated an overlap of 99.1 +/- 0:2%.
Outreach Material:
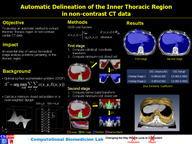

Associated Publications:
- D.R. Chittajallu, P. Balanša, and I.A. Kakadiaris, "Automatic delineation of the inner thoracic region in non-contrast CT data," in Proc. 31st International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Minneapolis, MN, Sep. 2-6 2009.
Abstract:
The diaphragm is a thin double-domed muscle that separates the thoracic and abdominal cavities. An accurate delineation of the diaphragm surface will be useful in providing a good region of interest for segmentation problems pertaining to the thoracic and abdominal cavities. In this paper, we present a fully automatic 3D graph-based method for the segmentation of the diaphragm in non-contrast CT data. In particular, we reformulate the diaphragm segmentation problem as an optimal surface segmentation problem in a volumetric graph. Comparison of the results obtained using our method with manual segmentations performed by an expert on non-contrast cardiac CT scans of 7 randomly selected patients indicated an overlap of 94.20 +/- 0:01%.
Outreach Material:

Associated Publications:
- R. Yalamanchili, D.R. Chittajallu, P. Balanša, B. Tamarappoo, D.S. Berman, D. Dey, and I.A. Kakadiaris. "Automated segmentation of the Diaphragm in non-contrast CT images," In Proc. IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI), Rotterdam, Netherlands, April 14-17, 2010.
Abstract:
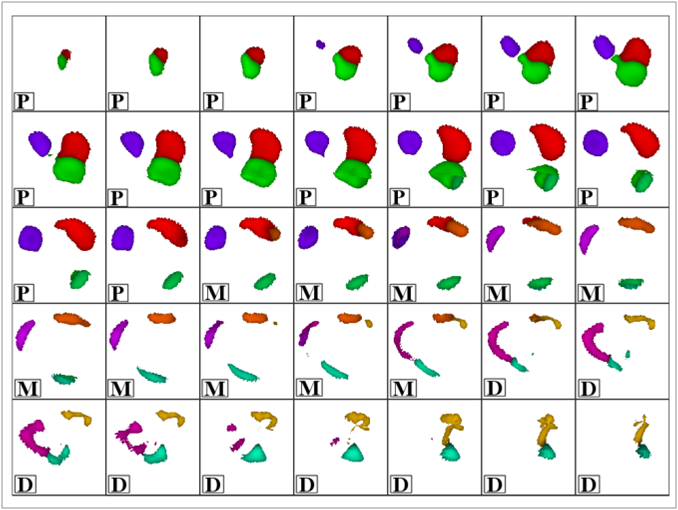
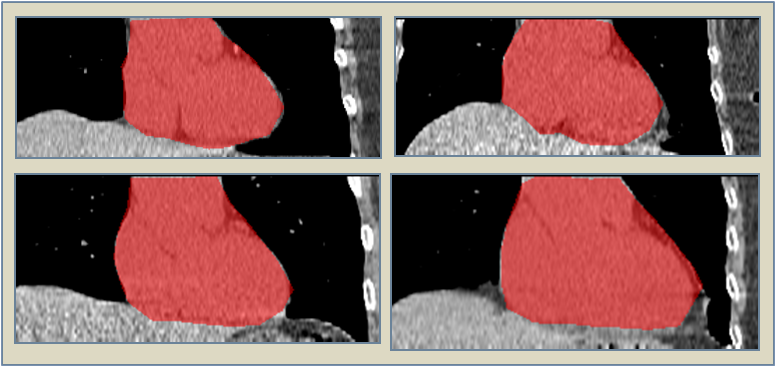
Image segmentation is, in general, an ill-posed problem and additional constraints need to be imposed in order to achieve the desired result. Particularly in the field of medical image segmentation, a significant amount of prior knowledge is available that can be used to constrain the solution space of the segmentation problem. However, most of this prior knowledge is, in general, vague or imprecise in nature, which makes it very difficult to model. This is the problem that is addressed here. Specifically, in this work, we present Fuzzy-Cuts, a novel, knowledge-driven, graph-based method for medical image segmentation. We cast the problem of image segmentation as the Maximum A Posteriori (MAP) estimation of a Markov Random Field (MRF) which, in essence, is equivalent to the minimization of the corresponding Gibbs energy function. Considering the inherent imprecision that is common in the a priori description of objects in medical images, we propose a fuzzy theoretic model to incorporate knowledge-driven constraints into the MAP-MRF formulation. In particular, we focus on prior information about the object's location, appearance and spatial connectivity to a known seed region inside the object. To that end, we introduce fuzzy connectivity and fuzzy location priors that are used in combination to define the first-order clique potential of the Gibbs energy function. In our experiments, we demonstrate the application of the proposed method to the challenging problem of heart segmentation in non-contrast computed tomography (CT) data.
Outreach Material:
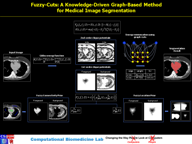
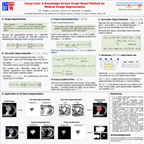
Associated Publications:
- D. R. Chittajallu, G. Brunner, U. Kurkure, R. Yalamanchili, and I. A. Kakadiaris, "Fuzzy-cuts: A knowledge-driven graph-based method for medical image segmentation," in Proc. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR) Miami Beach, FL, 2009.
Abstract:
In this work, we present a knowledge-driven Markov Random Field (MRF) model for the segmentation of organs in medical images with particular emphasis on the incorporation of shape constraints into the segmentation problem. We cast the problem of image segmentation as the Maximum A Posteriori (MAP) estimation of a Markov Random Field which, in essence, is equivalent to the minimization of the corresponding Gibbs energy function. We then incorporate a set of constraints into the Gibbs energy function that collectively force the resulting segmentation contour/surface to have a shape similar to that of a given shape template. In particular, we introduce a flux-maximization constraint and a generalized template-based star-shape constraint that are encoded into the first- and second-order clique potentials of the Gibbs energy function, respectively. Our main contribution is in the translation of a set of global notions about the shape of the desired segmentation contour into a set of local measures that can be conveniently encoded into the Gibbs energy function and used in combination with other traditionally used constraints derived from image information. In our experiments, we demonstrate the application of the proposed method to the challenging problem of heart segmentation in non-contrast computed tomography (CT) data.
Outreach Material:
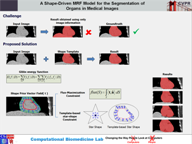
Associated Publications:
- D.R. Chittajallu, S. Shah, I.A. Kakadiaris, "A Shape-Driven MRF Model for the Segmentation of Organs in Medical Images," In Proc. IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), San Francisco, CA, 2010.
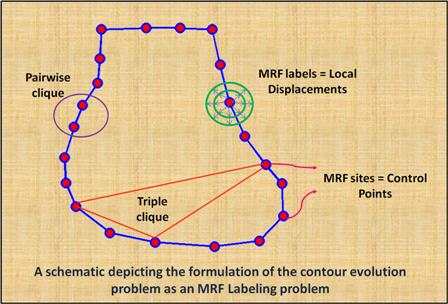
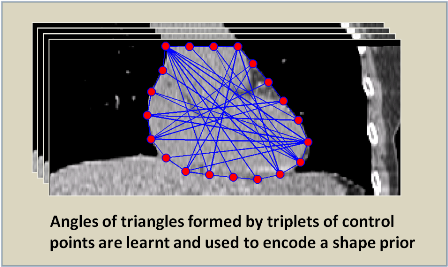
Abstract:
Image segmentation is, in general, an ill-posed problem and additional constraints need to be imposed in order to achieve the desired segmentation result. While segmenting organs in medical images, which is the topic of this paper, a significant amount of prior knowledge about the shape, appearance, and location of the organs is available that can be used to constrain the solution space of the segmentation problem. Among the various types of prior information, the incorporation of prior information about shape, in particular, is very challenging. In this work, we present an explicit shape-constrained MAP-MRF-based contour evolution method for the segmentation of organs in 2D medical images. Specifically, we represent the segmentation contour explicitly as a chain of control points. We then cast the segmentation problem as a contour evolution problem, wherein the evolution of the contour is performed by iteratively solving a MAP-MRF labeling problem. The evolution of the contour is governed by three types of prior information, namely: (i) appearance prior, (ii) boundary-edgeness prior, and (iii) shape prior, each of which are incorporated as clique potentials into the MAP-MRF problem. We use the master-slave dual decomposition framework to solve the MAP-MRF labeling problem in each iteration. In our experiments, we demonstrate the application of the proposed method to the challenging problem of heart segmentation in non-contrast computed tomography data.
Outreach Material:

Associated Publications:
- D.R. Chittajallu, N. Paragios, I.A. Kakadiaris, "An Explicit Shape-constrained MRF-based Contour Evolution Method for Medical Image Segmentation," IEEE Journal of Biomedical and Health Informatics (formely IEEE TITB), 2013 (In press).